Filtering by Retention Time#
The option “filter_by_rt” (-f) can be used to filter out regions of all chromatograms provided as an input. Not only will PyParse remove any peaks in these regions from consideration, but it will also recalculate all peak percentage areas to reflect the fact that this region was removed.
Usage#
To use this option, a range (or multiple ranges) should be provided in the form: “[first_retention_time]-[second_retention_time]”
For example, to filter out all peaks between 0 and 0.4 minutes, you would add the following argument:
-f 0-0.4
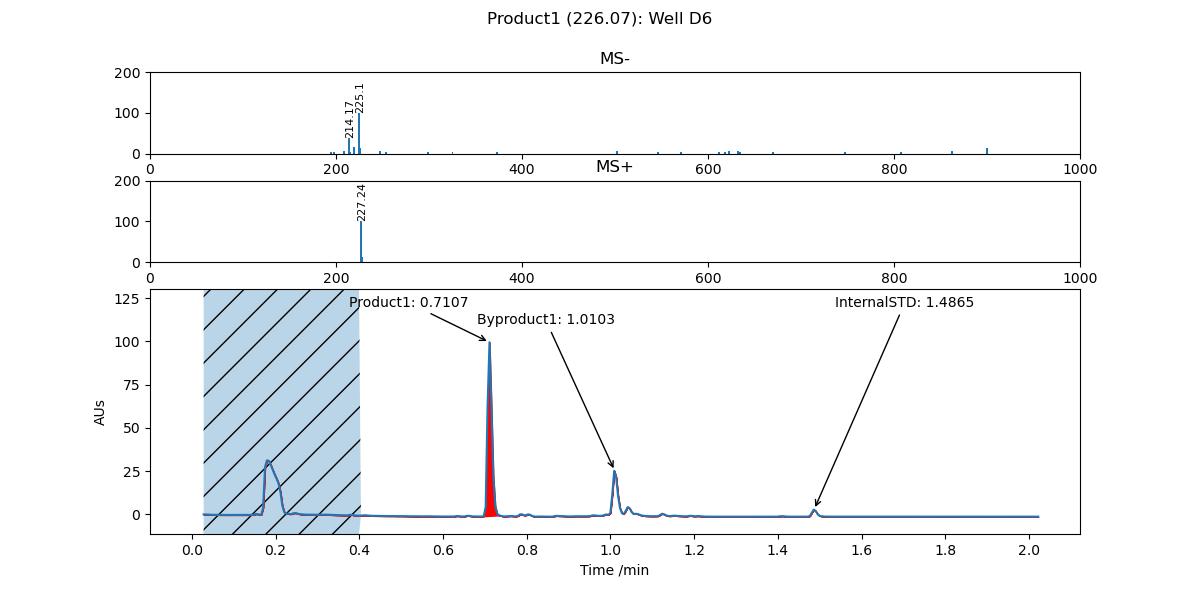
An example chromatogram where a specific range was filtered out.#
To filter out multiple ranges, input each range separated by a space:
-f 0-0.4 1.0-1.1
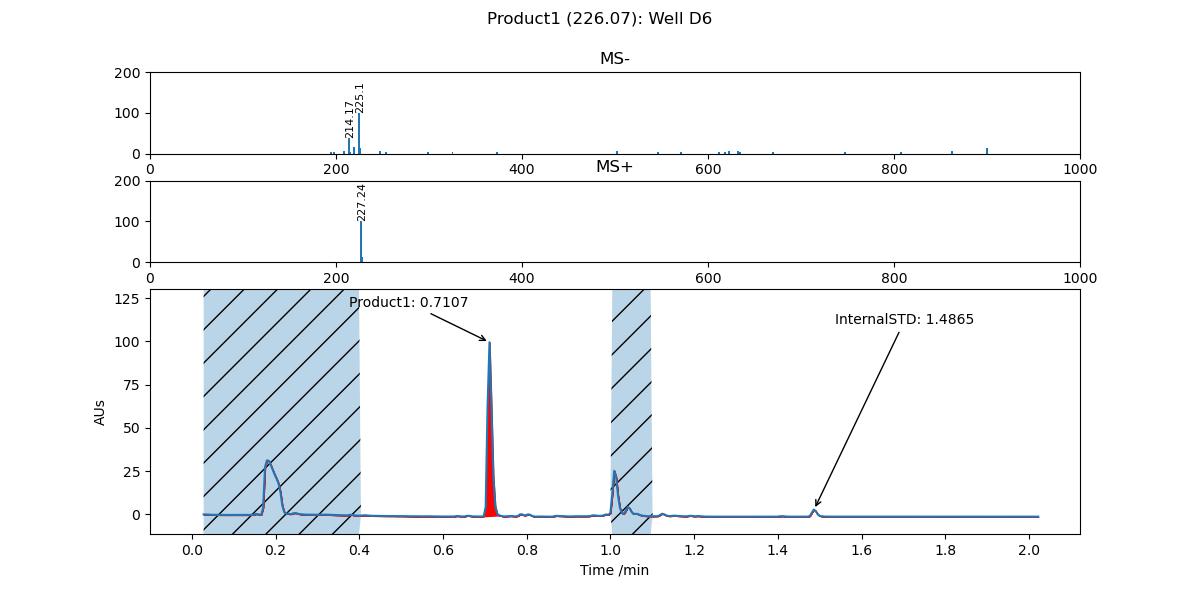
The result of this filtering is that the purity reported by PyParse is higher:


Top) Original purity value for Product1; Bottom) New purity value for Product1#
The purpose of this feature is to enable the user to remove solvent peaks (e.g. DMSO, toluene) from consideration. However, it should be noted that this feature should only be used with extreme caution to avoid misrepresenting the results!